
(From left) Doctoral student Hannah Yamagata, research assistant professor Kushol Gupta, and postdoctoral fellow Marshall Padilla holding 3D-printed models of nanoparticles.
(Image: Bella Ciervo)
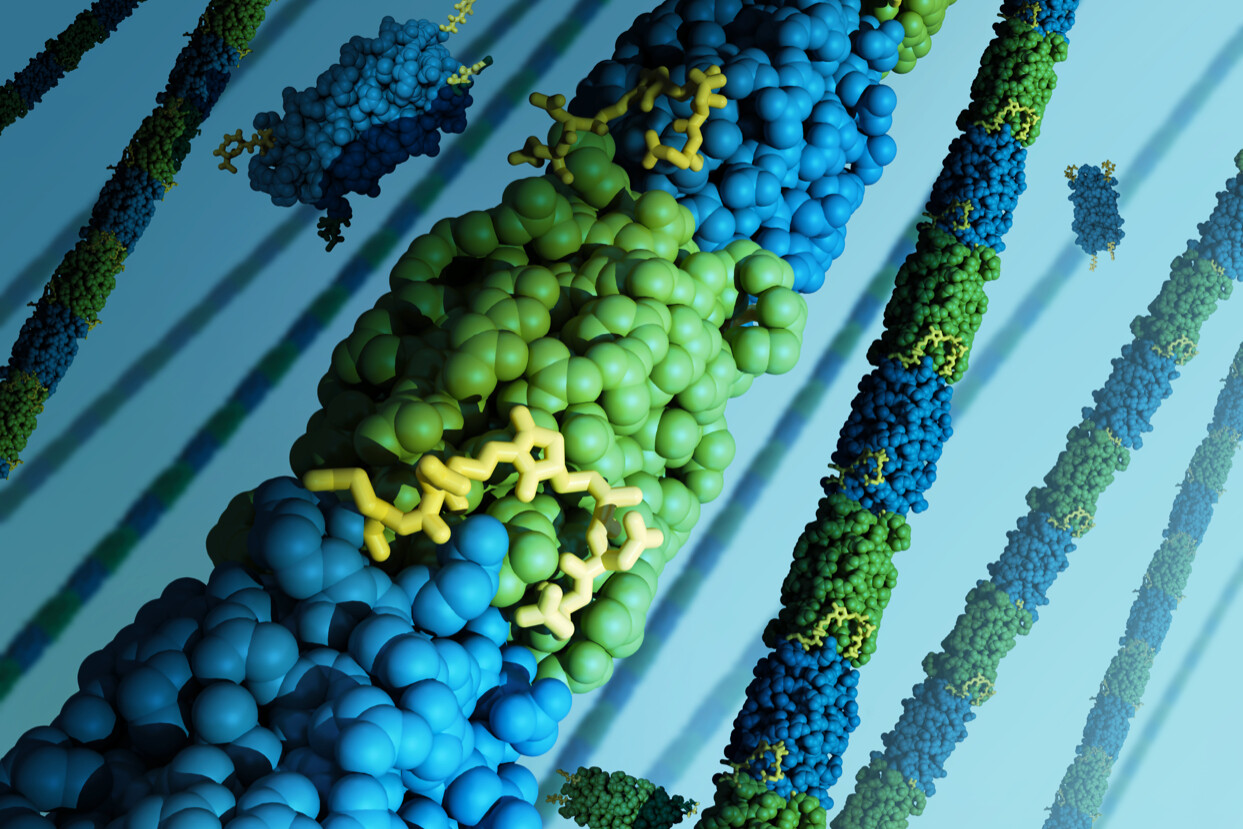
Polymers are materials that are made of repeating chains of smaller molecules, with natural examples including rubber and silk and manmade polymers including nylon, neoprene, plastics, and resins. Creating new polymers in the lab is a process that is difficult to control since structures often form as unorganized masses of intertwining threads, so researchers are looking for ways to create polymers that are easier to customize for a wider range of applications.
New research describes a novel type of synthetic polymer subunit that could usher in a new wave of applications. A collaboration between researchers at Penn and the University of Delaware shows how self-assembling and customizable polymers can be created by linking these new subunits, known as “bundlemers,” using efficient and controllable chemical reactions. The study was published in Nature.
Penn’s Jeffery Saven and Delaware’s Darrin Pochan have been collaborators since 2012, when they first began working together on designer materials. After developing several types of self-assembling chemical constructs with customizable features, they turned to Christopher Kloxin, also at Delaware, to test out some experimental methods they hoped could assemble new molecule combinations, in this case amino acids, into polymer building blocks.
Before the team synthesizes anything in the lab, Saven’s computational chemistry group uses models to analyze possible amino acid sequences to help the group decide which combinations would make the best candidates for synthesis and testing. Computational chemistry is an essential starting point due to the enormous number of sequences possible, with a choice of one of 20 natural amino acids at each position; even a short chain of 10 amino acids has more than 10 trillion possible combinations. Their models also take into account how amino acids interact with one another on a chain to provide insights into what types of structures, like helixes or rods, the sequences might form.
Saven’s group then works closely with the Delaware team to discuss other practical considerations that might not be captured by their models. “Some sequences might be easier to make than others, or you might run into an issue with the solubility that can be hard to anticipate from computation,” says Saven.
The resulting bundlemers, a term coined by the Delaware researchers, are made of four individual peptides, a term for any short chain of amino acids, that resemble nanoscale cylinders. The cylinders are then linked end to end through a highly efficient and controlled series of chemical reactions known as “click” chemistry. The type of linking reaction used can also help control the polymer’s flexibility.
What’s unique about some of the bundlemers they created is that they are incredibly rigid. Normally, polymers are loose and flexible, like cooked spaghetti, but certain bundlemers are long, thin, and strong, like spaghetti-thin rods of steel. The bundlemers have more stiffness per monomer weight than any known synthetic or natural polymer.
Saven says that it’s unusual for materials made of short, self-assembling peptides to form such long, thin, and straight objects and that the bundlemers’ overall structure doesn’t resemble anything seen in nature. “In nature, there are microtubules and actin filaments inside cells that support the cell’s architecture, but those tend to be much larger in terms of the filament radius and size of the building block,” he adds.
Beyond possible future applications ranging from high-performance fibers, biomedicines, or as substitutes for superstrong materials, the bundlemers can also become a platform for other applications not yet imagined. “These are tools for anybody to use, whether you’re a chemist, engineer, or physicist,” says Pochan. “It’s even hard to think of an equivalent material or experimental tool people use widely. It’s like a toolbox for anybody to design future things.”
The researchers are now planning to study the mechanism by which these thin and rigid structures form. “We’re now trying to figure out why that is and what some of the molecular determinants are,” says Saven, adding that computational chemistry will play an important role in predicting how chains of amino acids come together to encode specific structures. “Understanding how to build new compositions can be very hard to do. In that respect, computational chemistry and theoretical chemistry can be essential.”
Co-authors of this publication include the University of Delaware’s Jeffrey Caplan, Nicole Halaszynski, Christopher J. Kloxin, Jeeyoung Lee, Darrin J. Pochan, Nairiti Sinha, Bryan Sutherland, Yu Tian, and Dongdong Wu and Penn’s Jeffery G. Saven and Huixi Violet Zhang.
This research was supported by Department of Energy Office of Basic Energy Sciences, Biomolecular Materials Program grants DE-SC0019355 and DE-SC0019282; National Institute of Standards and Technology cooperative agreements 70NANB12H239 and 70NANB17H302; U.S. Department of Commerce, National Science Foundation Agreement DMR-0944772 and grants DMR-1120901, CHE-1709518; and National Institutes of Health RO1 EB006006, COBRE 1P30, GM110758, and 1P20 RR01771.
Portions of this text were adapted from a news release from the University of Delaware.
Jeffery Saven is a professor in the Department of Chemistry in the School of Arts and Sciences at the University of Pennsylvania.
Erica K. Brockmeier

(From left) Doctoral student Hannah Yamagata, research assistant professor Kushol Gupta, and postdoctoral fellow Marshall Padilla holding 3D-printed models of nanoparticles.
(Image: Bella Ciervo)

Jin Liu, Penn’s newest economics faculty member, specializes in international trade.
nocred

nocred

nocred